The data build tool (dbt) is an effective data transformation tool and it supports key AWS analytics services – Redshift, Glue, EMR and Athena. In the previous posts, we discussed benefits of a common data transformation tool and the potential of dbt to cover a wide range of data projects from data warehousing to data lake to data lakehouse. Demo data projects that target Redshift Serverless and Glue are illustrated as well. In part 3 of the dbt on AWS series, we discuss data transformation pipelines using dbt on Amazon EMR. Subsets of IMDb data are used as source and data models are developed in multiple layers according to the dbt best practices. A list of posts of this series can be found below.
Part 3 – EMR on EC2 (this post)
Below shows an overview diagram of the scope of this dbt on AWS series. EMR is highlighted as it is discussed in this post.
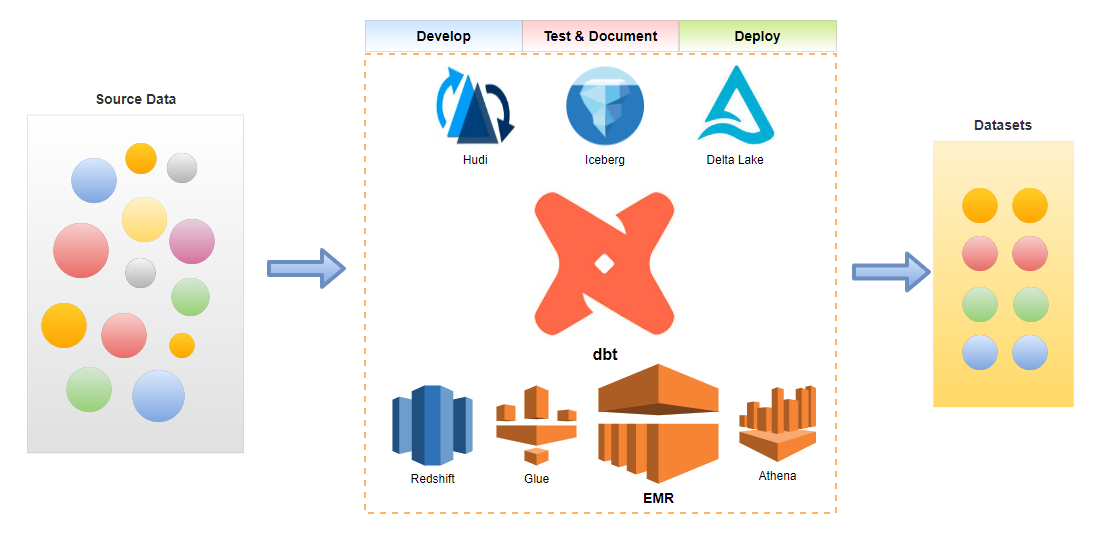
Infrastructure
The infrastructure hosting this solution leverages an Amazon EMR cluster and a S3 bucket. We also need a VPN server so that a developer can connect to the EMR cluster in a private subnet. It is extended from a previous post and the resources covered there (VPC, subnets, auto scaling group for VPN etc) are not repeated. All resources are deployed using Terraform and the source can be found in the GitHub repository of this post.
EMR Cluster
The EMR 6.7.0 release is deployed with single master and core node instances. It is configured to use the AWS Glue Data Catalog as the metastore for Hive and Spark SQL and it is done by adding the corresponding configuration classification. Also a managed scaling policy is created so that up to 4 additional task instances are added to the cluster. Note an additional security group is attached to the master and core groups for VPN access – the details of that security group is shown below.
# dbt-on-aws/emr-ec2/infra/emr.tf |
The following security group is created to enable access from the VPN server to the EMR instances. Note that the inbound rule is created only when the local.vpn.to_create variable value is true while the security group is created always – if the value is false, the security group has no inbound rule.
# dbt-on-aws/emr-ec2/infra/emr.tf |
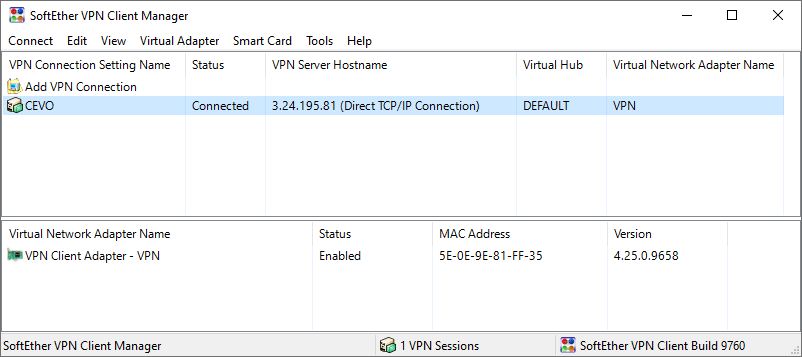
As in the previous post, we connect to the EMR cluster via SoftEther VPN. Instead of providing VPN related secrets as Terraform variables, they are created internally and stored to AWS Secrets Manager. The details can be found in dbt-on-aws/emr-ec2/infra/secrets.tf and the secret string can be retrieved as shown below.
$ aws secretsmanager get-secret-value –secret-id emr-ec2-all-secrets –query “SecretString” –output text |
The previous post demonstrates how to create a VPN user and to establish connection in detail. An example of a successful connection is shown below.
Glue Databases
We have two Glue databases. The source tables and the tables of the staging and intermediate layers are kept in the imdb database. The tables of the marts layer are stored in the imdb_analytics database.
# glue databases |
Project
We build a data transformation pipeline using subsets of IMDb data – seven titles and names related datasets are provided as gzipped, tab-separated-values (TSV) formatted files. The project ends up creating three tables that can be used for reporting and analysis.
Save Data to S3
The Axel download accelerator is used to download the data files locally followed by decompressing with the gzip utility. Note that simple retry logic is added as I see download failure from time to time. Finally the decompressed files are saved into the project S3 bucket using the S3 sync command.
# dbt-on-aws/emr-ec2/upload-data.sh |
Start Thrift JDBC/ODBC Server
The connection from dbt to the EMR cluster is made by the Thrift JDBC/ODBC server and it can be started by adding an EMR step as shown below.
$ cd emr-ec2 |
We can quickly check if the thrift server is started using the beeline JDBC client. The port is 10001 and, as connection is made in non-secure mode, we can simply enter the default username and a blank password. When we query databases, we see the Glue databases that are created earlier.
$ STACK_NAME=emr-ec2 … [hadoop@ip-10-0-113-195 ~]$ beeline |
Setup dbt Project
We use the dbt-spark adapter to work with the EMR cluster. As connection is made by the Thrift JDBC/ODBC server, it is necessary to install the adapter with the PyHive package. I use Ubuntu 20.04 in WSL 2 and it needs to install the libsasl2-dev apt package, which is required for one of the dependent packages of PyHive (pure-sasl). After installing it, we can install the dbt packages as usual.
$ sudo apt-get install libsasl2-dev |
We can initialise a dbt project with the dbt init command. We are required to specify project details – project name, host, connection method, port, schema and the number of threads. Note dbt creates the project profile to .dbt/profile.yml of the user home directory by default.
$ dbt init |
dbt initialises a project in a folder that matches to the project name and generates project boilerplate as shown below. Some of the main objects are dbt_project.yml, and the model folder. The former is required because dbt doesn’t know if a folder is a dbt project without it. Also it contains information that tells dbt how to operate on the project. The latter is for including dbt models, which is basically a set of SQL select statements. See dbt documentation for more details.
$ tree emr-ec2/emr_ec2/ -L 1 |
We can check connection to the EMR cluster with the dbt debug command as shown below.
$ dbt debug |
After initialisation, the model configuration is updated. The project materialisation is specified as view although it is the default materialisation. Also tags are added to the entire model folder as well as folders of specific layers – staging, intermediate and marts. As shown below, tags can simplify model execution.
# emr-ec2/emr_ec2/dbt_project.yml |
While we created source tables using Glue crawlers in part 2, they are created directly from S3 by the dbt_external_tables package in this post. Also the dbt_utils package is installed for adding tests to the final marts models. They can be installed by the dbt deps command.
# emr-ec2/emr_ec2/packages.yml packages: |
Create dbt Models
The models for this post are organised into three layers according to the dbt best practices – staging, intermediate and marts.
External Source
The seven tables that are loaded from S3 are dbt source tables and their details are declared in a YAML file (_imdb_sources.yml). Macros of the dbt_external_tables package parse properties of each table and execute SQL to create each of them. By doing so, we are able to refer to the source tables with the {{ source() }} function. Also we can add tests to source tables. For example two tests (unique, not_null) are added to the tconst column of the title_basics table below and these tests can be executed by the dbt test command.
# emr-ec2/emr_ec2/models/staging/imdb/_imdb__sources.yml version: 2 |
The source tables can be created by dbt run-operation stage_external_sources. Note that the following SQL is executed for the title_basics table under the hood.
create table imdb.title_basics ( |
Interestingly the header rows of the source tables are not skipped when they are queried by spark while they are skipped by Athena. They have to be filtered out in the stage models of the dbt project as spark is the query engine.
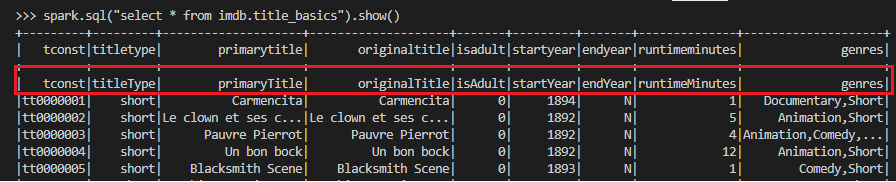
Staging
Based on the source tables, staging models are created. They are created as views, which is the project’s default materialisation. In the SQL statements, column names and data types are modified mainly.
# emr-ec2/emr_ec2/models/staging/imdb/stg_imdb__title_basics.sql with source as ( |
Below shows the file tree of the staging models. The staging models can be executed using the dbt run command. As we’ve added tags to the staging layer models, we can limit to execute only this layer by dbt run –select staging.
$ tree emr-ec2/emr_ec2/models/staging/ |
Note that the model materialisation of the staging and intermediate models is view and the dbt project creates VIRTUAL_VIEW tables. Although we are able to reference those tables in other models, they cannot be queried by Athena.
$ aws glue get-tables –database imdb \ |
Instead we can use spark sql to query the tables as shown below.
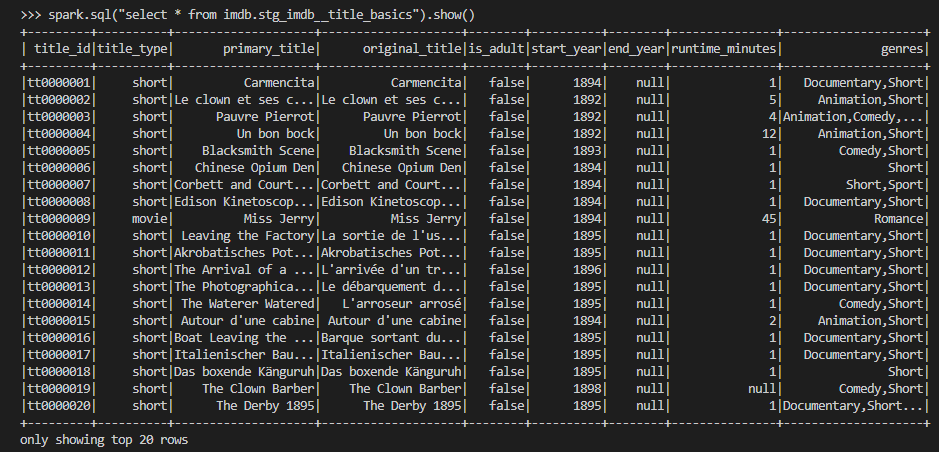
Intermediate
We can keep intermediate results in this layer so that the models of the final marts layer can be simplified. The source data includes columns where array values are kept as comma separated strings. For example, the genres column of the stg_imdb__title_basics model includes up to three genre values as shown in the previous screenshot. A total of seven columns in three models are columns of comma-separated strings and it is better to flatten them in the intermediate layer. Also, in order to avoid repetition, a dbt macro (flatten_fields) is created to share the column-flattening logic.
# emr-ec2/emr_ec2/macros/flatten_fields.sql {% macro flatten_fields(model, field_name, id_field_name) %} |
The macro function can be added inside a common table expression (CTE) by specifying the relevant model, field name to flatten and id field name.
— emr-ec2/emr_ec2/models/intermediate/title/int_genres_flattened_from_title_basics.sql with flattened as ( |
The intermediate models are also materialised as views and we can check the array columns are flattened as expected.
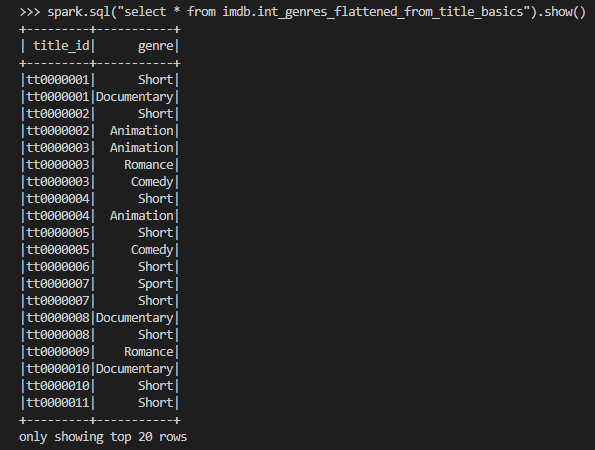
Below shows the file tree of the intermediate models. Similar to the staging models, the intermediate models can be executed by dbt run –select intermediate.
$ tree emr-ec2/emr_ec2/models/intermediate/ emr-ec2/emr_ec2/macros/ |
Marts
The models in the marts layer are configured to be materialised as tables in a custom schema. Their materialisation is set to table and the custom schema is specified as analytics while taking parquet as the file format. Note that the custom schema name becomes imdb_analytics according to the naming convention of dbt custom schemas. Models of both the staging and intermediate layers are used to create final models to be used for reporting and analytics.
— emr-ec2/emr_ec2/models/marts/analytics/titles.sql {{ |
The details of the three models can be found in a YAML file (_analytics__models.yml). We can add tests to models and below we see tests of row count matching to their corresponding staging models.
# emr-ec2/emr_ec2/models/marts/analytics/_analytics__models.yml version: 2 |
The models of the marts layer can be tested using the dbt test command as shown below.
$ dbt test –select marts |
Below shows the file tree of the marts models. As with the other layers, the marts models can be executed by dbt run –select marts.
$ tree emr-ec2/emr_ec2/models/marts/ |
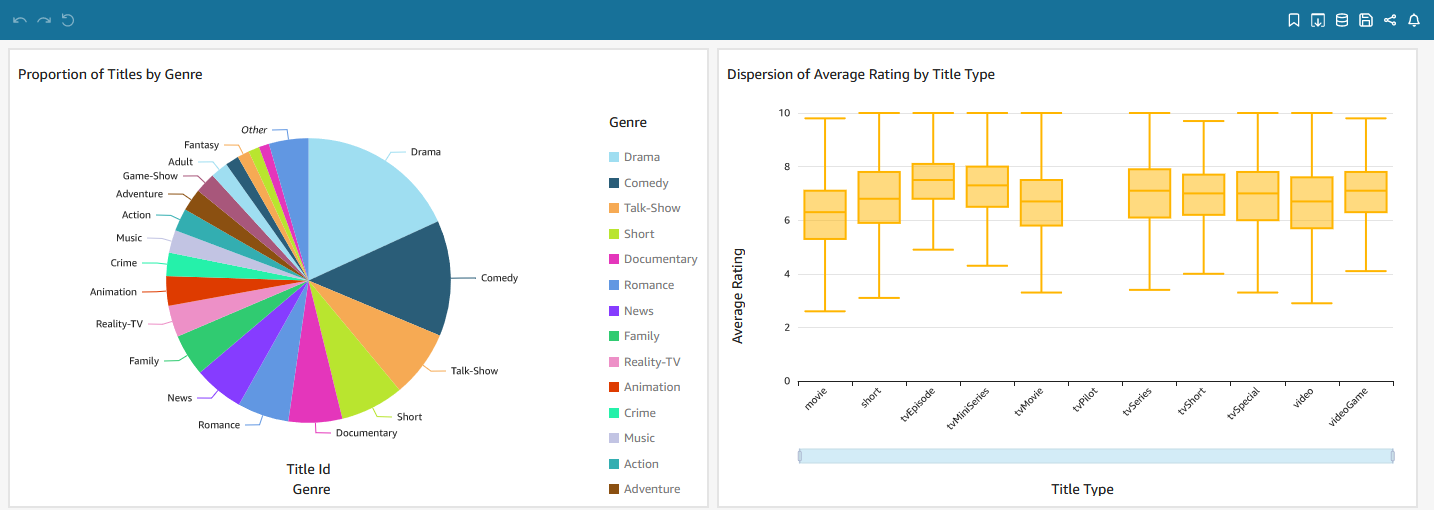
Build Dashboard
The models of the marts layer can be consumed by external tools such as Amazon QuickSight. Below shows an example dashboard. The pie chart on the left shows the proportion of titles by genre while the box plot on the right shows the dispersion of average rating by title type.
Generate dbt Documentation
A nice feature of dbt is documentation. It provides information about the project and the data warehouse and it facilitates consumers as well as other developers to discover and understand the datasets better. We can generate the project documents and start a document server as shown below.
$ dbt docs generate |
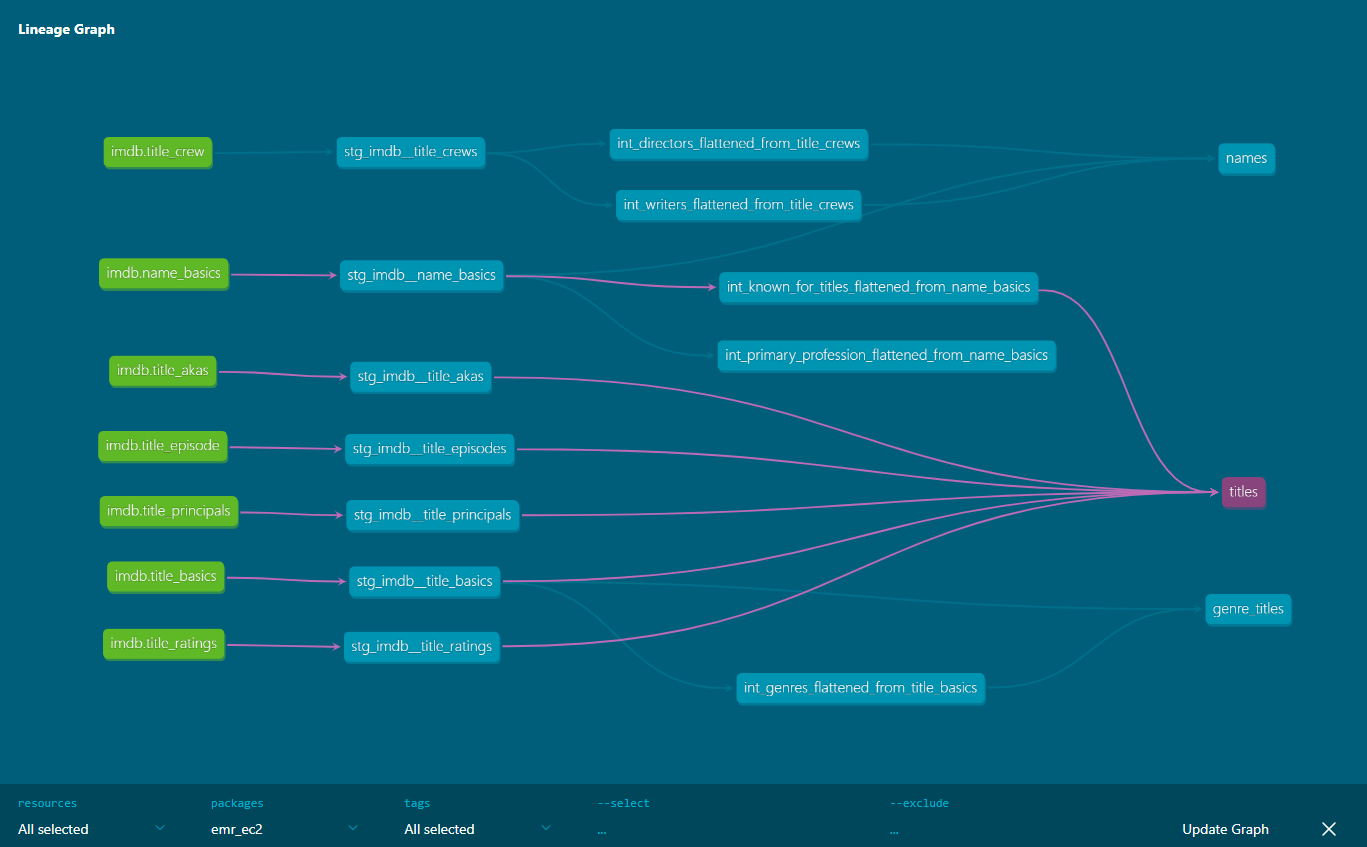
A very useful element of dbt documentation is data lineage, which provides an overall view about how data is transformed and consumed. Below we can see that the final titles model consumes all title-related stating models and an intermediate model from the name basics staging model.
Summary
In this post, we discussed how to build data transformation pipelines using dbt on Amazon EMR. Subsets of IMDb data are used as source and data models are developed in multiple layers according to the dbt best practices. dbt can be used as an effective tool for data transformation in a wide range of data projects from data warehousing to data lake to data lakehouse and it supports key AWS analytics services – Redshift, Glue, EMR and Athena. More examples of using dbt will be discussed in subsequent posts.




